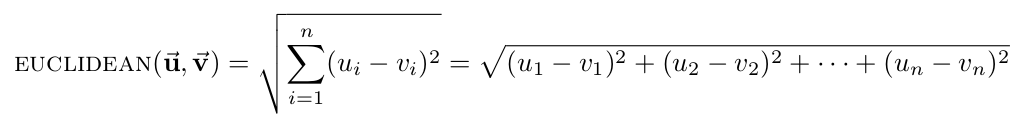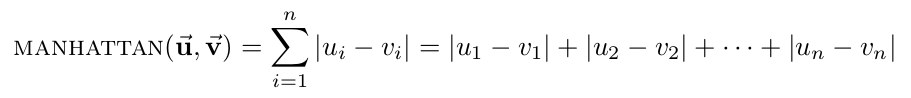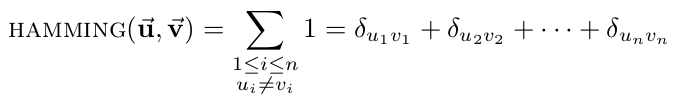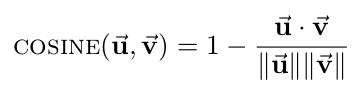Lab 6: Identification Trees
From 6.034 Wiki
Contents |
k-Nearest Neighbors
Multiple Choice: Drawing boundaries
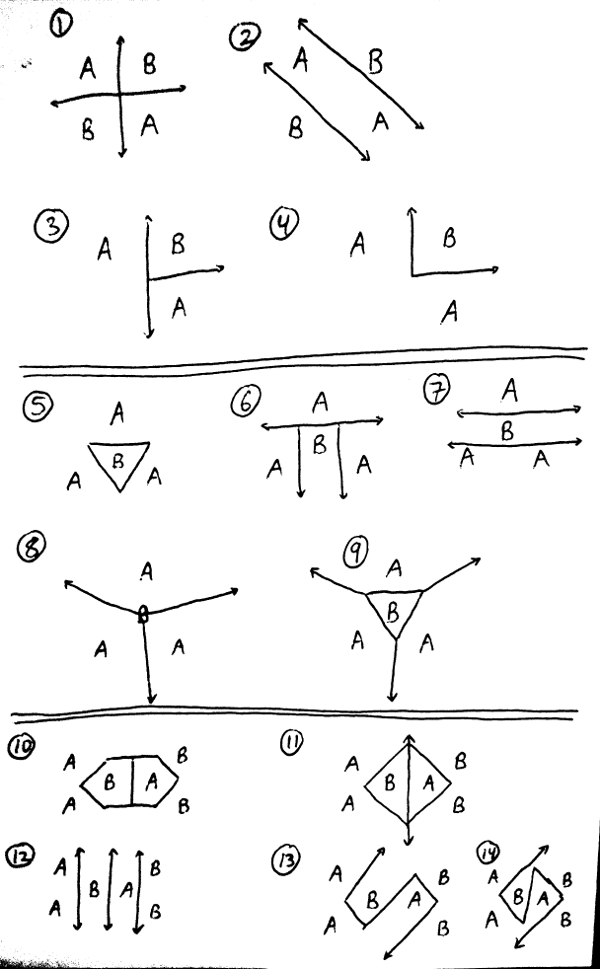
Below are sketches of 4 different data sets in Cartesian space, with numerous ways of drawing decision boundaries for each. For each sketch, answer the following question by filling in BOUNDARY_ANS_n (for n from 1 to 14) with an int 1, 2, 3, or 4:
- Which method could create the decision boundaries drawn in the sketch?
- Greedy, disorder-minimizing ID tree, using only tests of the form "X > threshold" or "Y > threshold"
- 1-nearest neighbors using Euclidean distance
- Both
- Neither
For explanations of the answers to some of these questions, search for "#BOUNDARY_ANS_n" in tests.py, for the appropriate value of n.
Python warm-up: Distance metrics
We can use many different distance metrics with k-nearest neighbors. In 6.034, we cover four such metrics:
- Euclidean distance: the straight-line distance between two points.
- Manhattan distance: the distance between two points assuming you can only move parallel to axes. In two dimensions, you can conceptualize this as only being able to move along the "grid lines."
- Hamming distance: the number of differing corresponding components between two vectors.
- Cosine distance: compares the angle between two vectors, where each point is represented as a vector from the origin to the point's coordinates. Note that cosine is a similarity measure, not a difference measure, because cos(0)=1 means two vectors are "perfectly similar," cos(90 deg)=0 represents two "perfectly orthogonal" vectors, and cos(180 deg)=-1 corresponds to two "maximally dissimilar" (or "perfectly opposite") vectors. For our code, however, we need this metric to be a difference measure so that smaller distances means two vectors are "more similar," to match all of the other distance metrics. Accordingly, we will define the cosine distance to be 1 minus the cosine of the angle between two vectors, so that our cosine distance varies between 0 (perfectly similar) and 2 (maximally dissimilar). Thus, the formula for cosine distance is:
It's also possible to transform data instead of using a different metric -- for instance, transforming to polar coordinates -- but in this lab we will only work with distance metrics in Cartesian coordinates.
We'll start with some basic functions for manipulating vectors, which may be useful for cosine_distance. For these, we will represent an n-dimensional vector as a list or tuple of n coordinates. dot_product should compute the dot product of two vectors, while norm computes the length of a vector. (There is a simple implementation of norm that uses dot_product.) Implement both functions:
def dot_product(u, v):
def norm(v):
Next, you'll implement each of the four distance metrics. For the rest of the k-nearest neighbors portion of this lab, we will represent data points as Point objects (described below in the API). Because Point objects are iterable (e.g. you can do for coord in point:), you should be able to call your vector methods on them directly. Each distance function should take in two Points and return the distance between them as a float or int. Implement each distance function:
def euclidean_distance(point1, point2):
def manhattan_distance(point1, point2):
def hamming_distance(point1, point2):
def cosine_distance(point1, point2):
Python hint: The built-in Python keyword zip may be useful. To learn more, type help(zip) in a Python command line, or read about it online.
Classifying Points with k-Nearest Neighbors
Now that we have defined four distance metrics, we will use them to classify points using k-nearest neighbors with various values of k.
First, write a function get_k_closest_points that takes in a point (a Point object), some data (a list of Point objects), a number k, and a distance metric (a function, such as euclidean_distance), and returns the k points in the data that are nearest to the input point, according to the distance metric. Break ties lexicographically using the points' coordinates (i.e. in case of a tie, sort by point.coords). This function can be written cleanly in 5 lines or less.
def get_k_closest_points(point, data, k, distance_metric):
A note about ties: There are two ways to get ties when classifying points using k-nearest neighbors.
- If two or more training points are equidistant to the test point, the "k nearest" points may be undefined. If the equidistant points have the same classification, it doesn't make a difference, but in some cases they could cause the classification to be ambiguous. In this lab, these cases are handled in get_k_closest_points by breaking ties lexicographically.
- If the two most likely classifications are equally represented among the k nearest training points, the classification is ambiguous. In other words, the k-nearest neighbors are voting on a classification, and if the vote is tied, there needs to be some mechanism for breaking the tie. In this lab, we will assume that there are no voting ties, so you don't need to worry about tie-breaking. (All of the test cases in this section are designed to ensure that there will not be any voting ties.)
Your task is to use that to write a function that classifies a point, given a set of data (as a list of Points), a value of k, and a distance metric (a function):
def knn_classify_point(point, data, k, distance_metric):
Python hint: To get the mode of a list, there's a neat one-line trick using max with the key argument, the list.count function, and converting a list to a set to get the unique elements. .count is a built-in list method that counts how many times an item occurs in a list. For example, [10, 20, 20, 30].count(20) -> 2.
Cross-validation: Choosing the best k and distance metric
There are many possible implementations of cross validation, but the general idea is to separate your data into a training set and a test set, then use the test set to determine how well the training parameters worked. For k-nearest neighbors, this means choosing a value of k and a distance metric, then testing each point in the test set to see whether they are classified correctly. The "cross" part of cross-validation comes from the idea that you can re-separate your data multiple times, so that different subsets of the data take turns being in the training and test sets, respectively.
For the next function, you'll implement a specific type of cross-validation known as leave-one-out cross-validation. If there are n points in the data set, you'll separate the data n times into a training set of size n-1 (all the points except one) and a test set of size one (the one point being left out). Implement the function cross_validate, which takes in a set of data, a value of k, and a distance metric, and returns the fraction of points that were classified correctly using leave-one-out cross-validation.
def cross_validate(data, k, distance_metric):
Use your cross-validation method to write a function that takes in some data (a list of Points) and tries every combination of reasonable values of k and the four distance metrics we defined above, then returns a tuple (k, distance_metric) representing the value of k and the distance metric that produce the best results with cross validation:
def find_best_k_and_metric(data):
More Multiple Choice
kNN Questions 1-3: More tree classification
These questions refer to the k-nearest neighbors data on classifying trees, from 2014 Quiz 2, Part B.
Recall the following terms:
- Overfitting: Occurs when small amounts of noise in the training data are treated as meaningful trends in the population. (i.e. when relying too heavily on the data, or overutilizing the data, causes a poor classification rate)
- Underfitting: Occurs when the algorithm blurs out differences in the training data that reflect genuine trends in the population. (i.e. when ignoring information in the data, or underutilizing the data, causes a poor classification rate)
kNN Question 1: Consider the results of cross-validation using k=1 and the Euclidean distance metric. Why is 1 not a good value for k? Answer "Overfitting" or "Underfitting" in kNN_ANSWER_1.
kNN Question 2: Consider the results of cross-validation using k=7 and the Euclidean distance metric. Why is 7 not a good value for k? Answer "Overfitting" or "Underfitting" in kNN_ANSWER_2.
kNN Question 3: Euclidean distance doesn't seem to do very well with any value of k (for this data). Which distance metric yields the highest classification rate for this dataset? Answer 1, 2, 3, or 4 in kNN_ANSWER_3:
- Euclidean distance
- Manhattan distance
- Hamming distance
- Cosine distance
kNN Questions 4-7: Choosing metrics
kNN Question 4: Suppose you wanted to classify a mysterious drink as coffee, energy drink, or soda based on the amount of caffeine and amount of sugar per 1-cup serving. Coffee typically contains a large amount of caffeine, lemonade typically contains a large amount of sugar, and energy drinks typically contain a large amount of both. Which distance metric would work best, and why? Answer 1, 2, 3, or 4 in kNN_ANSWER_4:
- Euclidean distance, because you're comparing standard numeric quantities
- Manhattan distance, because it makes sense to array the training data points in a grid
- Hamming distance, because the features "contains caffeine" and "contains sugar" are boolean values
- Cosine distance, because you care more about the ratio of caffeine to sugar than the actual amount
kNN Question 5: Suppose you add a fourth possible classification, water, which contains no caffeine or sugar. Now which distance metric would work best, and why? Answer 1, 2, 3, or 4 in kNN_ANSWER_5:
- Euclidean distance, because you're still comparing standard numeric quantities, and it's now more difficult to compare ratios
- Manhattan distance, because there are now four points, so it makes even more sense to array them in a grid
- Hamming distance, because the features "contains caffeine" and "contains sugar" are still boolean values
- Cosine distance, because you still care more about the ratio of caffeine to sugar than the actual amount
kNN Question 6: While visiting Manhattan, you discover that there are 3 different types of taxis, each of which has a distinctive logo. Each type of taxi comes in many makes and models of cars. Which distance metric would work best for classifying additional taxis based on their logos, makes, and models? Answer 1, 2, 3, or 4 in kNN_ANSWER_6:
- Euclidean distance, because there's a lot of variability in make and model
- Manhattan distance, because the streets meet at right angles (and because you're in Manhattan and classifying taxicabs)
- Hamming distance, because the features are non-numeric
- Cosine distance, because it will separate taxis by how different their makes and models are
kNN Question 7: Why might an ID tree work better for classifying the taxis from Question 6? Answer 1, 2, 3, or 4 in kNN_ANSWER_7:
- k-nearest neighbors requires data to be graphed on a coordinate system, but the training data isn't quantitative
- Some taxis may have the same make and model, and k-nearest neighbors can't handle identical training points
- An ID tree can ignore irrelevant features, such as make and model
- Taxis aren't guaranteed to stay within their neighborhood in Manhattan
For explanations of the answers to some of these questions, search for "#kNN_ANSWER_n" in tests.py, for the appropriate value of n.
API Reference Documentation
In lab5.py, we have defined for you...
- log2: a function representing the logarithm to base 2, taking in a single number and returning log2 of the number.
- INF: a constant representing positive infinity, as in previous labs
In addition, the file api.py defines the Classifier and IdentificationTreeNode classes, as well as some helper functions for creating Classifiers, all described below.
Classifier
Classifier objects are used for constructing and manipulating ID trees.
A Classifier has one attribute and one method:
- classifier.name, the name of the classifier.
- classifier.classify(point), which takes in a point and returns its classification. (A classification can be a string (e.g. "Vampire"), a boolean value (True or False) or an int.)
In our ID trees, a point is represented as a dict mapping feature names to their values. For example:
point = {"X": 1, "Y": 2, "Zombie": True}
You can create new Classifiers using these provided methods:
- feature_test(key): Takes in the name of a feature (such as "X" or "Accent" or "Height"). Returns a new Classifier that classifies based on the value of that feature.
- threshold_test(key, threshold): Takes in the name of a numeric feature and a threshold value. Returns a new binary Classifier that performs the test point[key] > threshold and returns "Yes" or "No" for each point.
For example:
feature_test_X = feature_test("X")
feature_test_X_threshold = threshold_test("X", 6)
Or you can create your own Classifier using the Classifier constructor, for example:
feature_test_vector_length = Classifier("vector_length", lambda pt: (pt.get("X")**2 + pt.get("Y")**2)**0.5)
IdentificationTreeNode
In this lab, an ID tree is represented recursively as a tree of IdentificationTreeNode objects. In particular, an IdentificationTreeNode object fully represents an entire ID tree rooted at that node.
For example, suppose we have an IdentificationTreeNode called id_tree_node Then, id_tree_node's children are themselves IdentificationTreeNode objects, each fully describing the sub-trees of id_tree_node. However, if id_tree_node has no children, then id_tree_node is a leaf, meaning that it represents a homogeneous (by classification) set of data points. Furthermore, any datum at this node is definitively classified by that leaf's classification.
As such, in a completed ID tree, each node is either
- a leaf node with a classification such as "Vampire" or "Not Vampire" in the vampires example; or
- a non-leaf node with a classifier (such as "Accent" in the vampires example), with branches (one per classifier result, e.g. "Heavy", "Odd", and "None") leading to child nodes.
An IdentificationTreeNode has the following attributes and methods:
- id_tree_node.target_classifier, the single Classifier by which the tree classifies points (e.g. a Classifier representing "Vampire?" for the vampires example, or "Classification" for the angel data).
- id_tree_node.get_parent_branch_name(): returns the name of the branch leading to this node, or None if this is a root node.
- id_tree_node.is_leaf(): returns True if the node is a leaf (has a classification), otherwise False.
- id_tree_node.set_node_classification(classification): Sets this node's classification, thus defining this node as a leaf node. Modifies and returns the original id_tree_node. May print warnings if the node already has branches defined.
- id_tree_node.get_node_classification(): returns this node's classification, if it has one. Otherwise, returns None.
- id_tree_node.set_classifier_and_expand(classifier, features): Takes in a Classifier object and a list of features (e.g. ["Heavy", "Odd", "None"]), then uses the Classifier and list of features to set the current node's classifier and add branches below the current node, instantiating a new child node for each branch. Modifies and returns the original id_tree_node. May print warnings if the specified Classifier is inadvisable. (For your convenience, features can also be a dictionary whose keys are feature names.)
- id_tree_node.get_classifier(): returns the classifier associated with this node, if it has one. Otherwise, returns None.
- id_tree_node.apply_classifier(point): Applies this node's classifier to a given data point by following the appropriate branch of the tree, then returns the child node. If this node is a leaf node (and thus doesn't have a classifier), raises a ClassifierError.
- id_tree_node.get_branches(): returns a dictionary mapping this node's branch names to its child nodes (e.g. {'Heavy':<IdentificationTreeNode object>, 'Odd': <IdentificationTreeNode object>, 'None': <IdentificationTreeNode object>})
- id_tree_node.copy(): Returns a (deep) copy of the node
- id_tree_node.print_with_data(data): Given a list of points, automatically classifies each point and prints a graphical representation of the ID tree with each point at the appropriate node.
The constructor for instantiating a new IdentificationTreeNode requires 1-2 arguments:
- target_classifier: The Classifier by which the overall tree should classify points
- parent_branch_name (optional): The name of the branch leading into this node, if the node is not the root node.
For example:
vampire_tree_root_node = IdentificationTreeNode(feature_test("Vampire?"))
another_node = IdentificationTreeNode(feature_test("Vampire?"), "Odd")
Appendix
How to handle exceptions
(using NoGoodClassifiersError as an example)
| Python command | Effect |
|---|---|
| raise NoGoodClassifiersError("message") | Stops execution by raising, or throwing, a NoGoodClassifiersError with the specified error message |
| try: #code here | Defines the beginning of a block of code that might raise an exception |
| except NoGoodClassifiersError: #code here | Follows a try block. Defines what to do if a NoGoodClassifiersError was raised in the try block. |